A High‐Throughput Screening Method for Identification of Inhibitors of the Deubiquitinating Enzyme USP14
互联网
- Abstract
- Table of Contents
- Materials
- Figures
- Literature Cited
Abstract
Deubiquitinating enzymes (DUBs) reverse the process of ubiquitination, and number nearly 100 in humans. In principle, DUBs represent promising drug targets, as several of the enzymes have been implicated in human diseases. The isopeptidase activity of DUBs can be selectively inhibited by targeting the catalytic site with drug?like compounds. Notably, the mammalian 26S proteasome is associated with three major DUBs: RPN11, UCH37, and USP14. Because the ubiquitin ?chain?trimming? activity of USP14 can inhibit proteasome function, inhibitors of USP14 can stimulate proteasomal degradation. We recently established a high?throughput screening (HTS) method to identify small?molecule inhibitors specific for USP14. The protocols in this article cover the necessary procedures for preparing assay reagents, performing HTS for USP14 inhibitors, and carrying out post?HTS analysis. Curr. Protoc. Chem. Biol. 4:311?330 © 2012 by John Wiley & Sons, Inc.
Keywords: 26S proteasome; USP14; deubiquitinating enzyme; high?throughput screening; small?molecule inhibitor
Table of Contents
- Introduction
- Basic Protocol 1: Measurement of Deubiquitination Activity of USP14
- Basic Protocol 2: High‐Throughput Screening of Small‐Molecule Inhibitors of Proteasome‐Associated USP14
- Basic Protocol 3: Secondary Screening and Analysis of Primary Hit Compounds of Proteasome‐Associated USP14
- Support Protocol 1: Purification of Human 26S Proteasomes that Lack Endogenous USP14 and are Devoid of UB‐AMC Hydrolysis Activity
- Support Protocol 2: Purification of Recombinant Human USP14 from E. coli
- Support Protocol 3: Synthesis and Purification of Ubiquitin‐AMC
- Reagents and Solutions
- Commentary
- Literature Cited
- Figures
Materials
Basic Protocol 1: Measurement of Deubiquitination Activity of USP14
Materials
Basic Protocol 2: High‐Throughput Screening of Small‐Molecule Inhibitors of Proteasome‐Associated USP14
Materials
Basic Protocol 3: Secondary Screening and Analysis of Primary Hit Compounds of Proteasome‐Associated USP14
Materials
Support Protocol 1: Purification of Human 26S Proteasomes that Lack Endogenous USP14 and are Devoid of UB‐AMC Hydrolysis Activity
Materials
Support Protocol 2: Purification of Recombinant Human USP14 from E. coli
Materials
Support Protocol 3: Synthesis and Purification of Ubiquitin‐AMC
Materials
|
Figures
-
Figure 1. DUB assays with Ub‐VS‐untreated 26 proteasome (A , 26S) or Ub‐VS‐treated 26 proteasome (B , VS‐26S) in the USP14 reconstitution system. Ub‐VS is a commercially available active‐site‐directed inhibitor that irreversibly inactivates thiol protease DUBs. 26S (1 nM) or VS‐26S (1 nM) in the absence or presence of indicated concentrations of recombinant USP14 wild‐type (WT) or catalytic inactive mutant (CA) was analyzed for the Ub‐AMC hydrolysis in a given time. Ub‐AMC (1 µM) was added as substrate. Note that most of the DUB activity of 26S in (A) comes from proteasome‐associated UCH37 since RPN11 does not cleave Ub‐AMC. RFU indicates relative fluorescence unit. Detailed descriptions for the assay reagents and the DUB assay can be found in the protocols. View Image -
Figure 2. Workflow of the HTS for small‐molecule inhibitors of proteasome‐associated USP14. The screening procedures were based on the equipment at the ICCB‐Longwood Screening Facility, Harvard Medical School (http://iccb.med.harvard.edu). View Image -
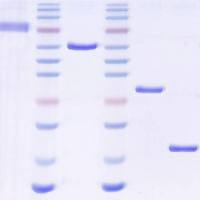
Figure 3. Representative preparations of VS‐26S (A ), Ub‐AMC (B ), and recombinant USP14 (C ). (A ) Native gel analysis of 26S and VS‐26S. The proteasome (7 µg) was analyzed for in‐gel peptidase activity with LLVY‐AMC as the substrate (left) or for Coomassie Brilliant Blue staining (middle, CBB) on a 3.6% native gel. SDS‐PAGE analysis of 7 µg of purified proteasome is shown (right, CBB). (B ) SDS‐PAGE analysis and CBB staining of purified Ub thiol esters (2 µg), and 1 µg of purified Ub‐AMC and HA‐tagged Ub‐AMC (HA‐Ub‐AMC). Free Ub (1 µg) was loaded for comparison. (C ) SDS‐PAGE analysis and CBB staining of purified USP14 WT and CA. A known amount of BSA was loaded for comparison. RP2 CP indicates doubly capped proteasome holoenzyme. View Image -
Figure 4. Schematic flowchart of the required amount of assay reagents for a given scale of screen. View Image
Videos
Literature Cited
| Altun, M., Kramer, H.B., Willems, L.I., McDermott, J.L., Leach, C.A., Goldenberg, S.J., Kumar, K.G., Konietzny, R., Fischer, R., Kogan, E., Mackeen, M.M., McGouran, J., Khoronenkova, S.V., Parsons, J.L., Dianov, G.L., Nicholson, B., and Kessler, B.M. 2011. Activity‐based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chem. Biol. 18:1401‐1412. | |
| Chen, J., Dexheimer, T.S., Ai, Y., Liang, Q., Villamil, M.A., Inglese, J., Maloney, D.J., Jadhav, A., Simeonov, A., and Zhuang, Z. 2011. Selective and cell‐active inhibitors of the USP1/UAF1 deubiquitinase complex reverse cisplatin resistance in non‐small cell lung cancer cells. Chem. Biol. 18:1390‐1400. | |
| Cohen, P. and Tcherpakov, M. 2010. Will the ubiquitin system furnish as many drug targets as protein kinases? Cell 143:686‐693. | |
| Colland, F. 2010. The therapeutic potential of deubiquitinating enzyme inhibitors. Biochem. Soc. Trans. 38:137‐143. | |
| D'Arcy, P., Brnjic, S., Olofsson, M.H., Fryknäs, M., Lindsten, K., Cesare, M.D., Perego, P., Sadeghi, B., Hassan, M., Larsson, R., and Linder, S. 2011. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat. Med. 17:1636‐1641. | |
| Elsasser, S., Schmidt, M., and Finley, D. 2005. Characterization of the proteasome using native gel electrophoresis. Methods Enzymol. 398:353‐363. | |
| Finley, D. 2009. Recognition and processing of ubiquitin‐protein conjugates by the proteasome. Annu. Rev. Biochem. 78:477‐513. | |
| Inglese, J., Shamu, C.E., and Guy, R.K. 2007a. Reporting data from high‐throughput screening of small‐molecule libraries. Nat. Chem. Biol. 3:438‐441. | |
| Inglese, J., Johnson, R.L., Simeonov, A., Xia, M., Zheng, W., Austin, C.P., and Auld, D.S. 2007b. High‐throughput screening assays for the identification of chemical probes. Nat. Chem. Biol. 3:466‐479. | |
| Lee, B.H., Lee, M.J., Park, S., Oh, D.C., Elsasser, S., Chen, P.C., Gartner, C., Dimova, N., Hanna, J., Gygi, S.P., Wilson, S.M., King, R.W., and Finley, D. 2010. Enhancement of proteasome activity by a small‐molecule inhibitor of USP14. Nature 467:179‐184. | |
| Lee, M.J., Lee, B.H., Hanna, J., King, R.W., and Finley, D. 2011. Trimming of ubiquitin chains by proteasome‐associated deubiquitinating enzymes. Mol. Cell. Proteomics 10:R110.003871. | |
| Liu, Y., Lashuel, H.A., Choi, S., Xing, X., Case, A., Ni, J., Yeh, L.A., Cuny, G.D., Stein, R.L., and Lansbury, P.T. 2003. Discovery of inhibitors that elucidate the role of UCH‐L1 activity in the H1299 lung cancer cell line. Chem. Biol. 10:837‐846. | |
| Malo, N., Hanley, J., Cerquozzi, S., Pelletier, J., and Nadon, R. 2006. Statistical practice in high‐throughput screening data analysis. Nat. Biotechnol. 24:167‐175. | |
| Ovaa, H., Galardy, P.J., and Ploegh, H.L. 2005. Mechanism‐based proteomics tools based on ubiquitin and ubiquitin‐like proteins: Synthesis of active site‐directed probes. Methods Enzymol. 399:468‐478. | |
| Reyes‐Turcu, F.E., Ventii, K.H., and Wilkinson, K.D. 2009. Regulation and cellular roles of ubiquitin‐specific deubiquitinating enzymes. Annu. Rev. Biochem. 78:363‐397. | |
| Rudnicki, S. and Johnston, S. 2009. Overview of liquid handling instrumentation for high‐throughput screening applications. Curr. Protoc. Chem. Biol. 1:43‐54. | |
| Sacco, J.J., Coulson, J.M., Clague, M.J., and Urbe', S. 2010. Emerging roles of deubiquitinases in cancer‐associated pathways. IUBMB Life 62:140‐157. | |
| Seidman, C.E., Struhl, K., Sheen, J., and Jessen, T. 1997. Introduction of plasmid DNA into cells. Curr. Protoc. Mol. Biol. 37:1.8.1‐1.8.10. | |
| Simonian, M.H. and Smith, J.A. 2006. Spectrophotometric and colorimetric determination of protein concentration. Curr. Protoc. Mol. Biol. 76:10.1.1‐10.1A.9. | |
| Wang, X., Chen, C.F., Baker, P.R., Chen, P.I., Kaiser, P., and Huang, L. 2007. Mass spectrometric characterization of the affinity‐purified human 26S proteasome complex. Biochemistry 46:3553‐3565. | |
| Wilkinson, KD, Gan‐Erdenem, T, Kolli, N. 2005. Derivitization of the C‐terminus of ubiquitin and ubiquitin‐like proteins using intein chemistry: Methods and uses. Methods Enzymol. 399:37‐51. | |
| Zhang, J.H., Chung, T.D., and Oldenburg, K.R. 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Scr. 4:67‐73. | |
| Zeng, X., Sigoillot, F., Gaur, S., Choi, S., Pfaff, K.L., Oh, D.C., Hathaway, N., Dimova, N., Cuny, G.D., and King, R.W. 2010. Pharmacologic inhibition of the anaphase‐promoting complex induces a spindle checkpoint‐dependent mitotic arrest in the absence of spindle damage. Cancer Cell 18:382‐395. | |
| Internet Resources | |
| http://iccb.med.harvard.edu | |
| The Web page of the ICCB screening facility at Harvard Medical School. This site provides general guidance and detailed information of small‐molecule inhibitor screening. | |
| http://www.ncbi.nlm.nih.gov/books/NBK53196/ | |
| Assay Guidance Manual by Eli Lilly & Company and the National Center for Advancing Translational Sciences at the NIH. |